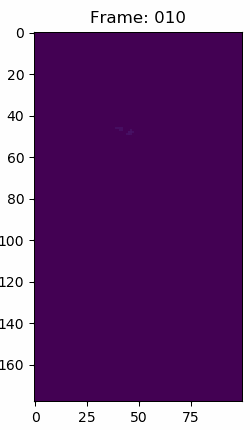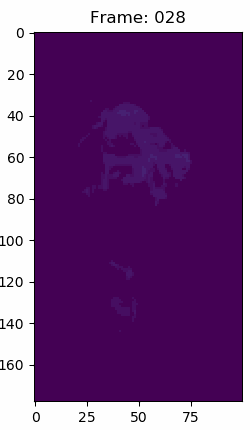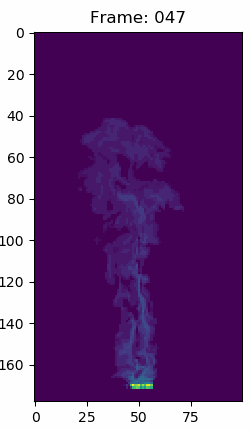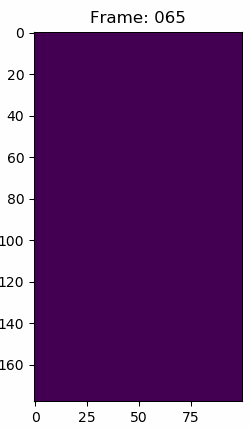
For context about the float data in the volumes. This is what simulation number 70 snapshot 150 looks like in matplotlib with a viridis color map, but the data in each voxel are simple float64 values. The other snapshops and simulations look similar.
This is how you load a single npz file in python, if you want to try it with the dataset. It just generates a generic numpy float array. So you could just make a tiny numpy array filled with random data yourself for the purpose of testing like this np.random.rand(3,3,3).
import numpy as np def npz_to_nparray(filepath): """ loads npz file from location :param filepath: string to file location :return: numpy array """ data = np.load(filepath) data = data[data.files[0]] # normal strange operations to get the the actual array inside data = np.transpose(data, (3, 1, 0, 2)) # turning the inner array layer outside and rightside up return data[0]
I have tried:
-I can load the npz files in python and in Houdini's python shell as a numpy array. I dont know how to put the data into a Houdini volume or which file format Houdini needs to import it.
-I tried to translate the npz files to vdb with the pyopenvdb libary for python. But after multiple days of trying I am unable to import pyopenvdb. I build pyopenvdb library on windows with vcpkg and cmake, but I cannot import the build package. I setup an ubuntu VM and tried importing pyopenvdb there, but I can not import it.
tl;dr: How to import animated smoke voxel data that is currently stored as a 4D python array?